Practicing Predictive Analytics using “R”
February 7, 2016 Leave a comment
I spent a Sunday on this code to answer some questions for a Coursera course. At this time this code is the norm in more than one such course. So I am just building muscle memory. I type this code and look at the result and learn what I learnt earlier.
If I don’t remember how to solve it I search but the point is that I have to be constantly in touch with “R” as well the fundamentals. My day job doesn’t let me do this. The other option is a book on Machine Learning like the one by Tom Mitchell but that takes foreover.
setwd("~/Documents/PredictiveAnalytics")
library(dplyr)
library(ggplot2)
library(rpart)
library(tree)
library(randomForest)
library(e1071)
library(caret)
seaflow <- read.csv(file="seaflow_21min.csv",head=TRUE)
final <-filter(seaflow, pop == "synecho")
print(nrow(final))
print( summary(seaflow))
print ( nrow(seaflow))
print( head(seaflow))
set.seed(555)
trainIndex <- createDataPartition( seaflow$file_id, p = 0.5, list=FALSE, times=1)
train <- seaflow[ trainIndex,]
test <- seaflow[ -trainIndex,]
print(mean(train$time))
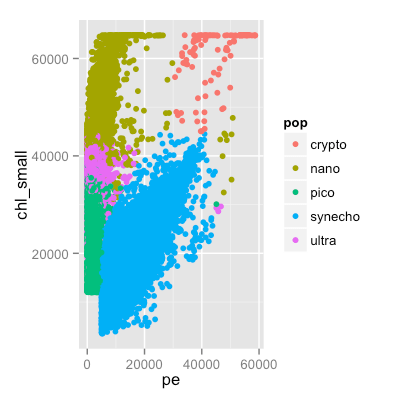
p <- ggplot( seaflow, aes( pe, chl_small, color = pop)) + geom_point()
dev.new(width=15, height=14)
print(p)
ggsave("~/predictiveanalytics.png", width=4, height=4, dpi=100)
fol <- formula(pop ~ fsc_small + fsc_perp + fsc_big + pe + chl_big + chl_small)
model <- rpart(fol, method="class", data=train)
print(model)
#plot(model)
#text(model, use.n = TRUE, all=TRUE, cex=0.9)
testprediction <- predict( model, newdata=test, type="class")
comparisonofpredictions <- testprediction == test$pop
accuracy <- sum(comparisonofpredictions) / length(comparisonofpredictions)
print( accuracy )
randomforestmodel <- randomForest( fol, data = train)
print(randomforestmodel)
testpredictionusingrandomforest <- predict( randomforestmodel, newdata=test, type="class")
comparisonofpredictions <- testpredictionusingrandomforest == test$pop
accuracy <- sum(comparisonofpredictions) / length(comparisonofpredictions)
print( accuracy )
print(importance(randomforestmodel))
svmmodel <- svm( fol, data = train)
testpredictionusingsvm <- predict( svmmodel, newdata=test, type="class")
comparisonofpredictions <- testpredictionusingsvm == test$pop
accuracy <- sum(comparisonofpredictions) / length(comparisonofpredictions)
print( accuracy )